CONP Portal | Dataset
Multicenter Single Subject Human MRI Phantom
| Dimensions: | resting BOLD, fieldmap BOLD, fieldmap DWI, DWI 65 directions, DWI 25 directions, T1W, T2W |
|---|---|
| Is About: | Homo sapiens, Homo sapiens, adult |
| Spatial Coverage: | North America |
| Other Dates: | Start Date: 2008-04-25 00:00:00 -- End Date: 2019-04-15 00:00:00 |
Description:
Dataset README information
Multicenter Single Subject Human MRI Phantom Dataset
Overview
This dataset consists of brain scans of a single human phantom acquired on multiple MRI devices
across North America over a period of 11 years. In addition to the human brain images, lego phantom
scans have been acquired in parallel for quality assessments over time across sites.
MRI modalities include: T1-weighted (t1w), T2-weighted (t2w), resting state BOLD (bold),
25-direction DWI (dwi25), 65-direction DWI (dwi65) as well as fieldmaps for the BOLD and
DWI acquisitions (fieldmapBOLD and fieldmapDWI).
The human phantom data were collected once a year during two MRI sessions. Those two sessions were either performed the same day or with a few days apart.
Data organization and file format
Data are organized by subject_ID/Visit_Label.
.
|__ subject-1
|__ visit-label-1
|__ images
|__ images.json
|__ phantom_<subject-1>_<visit-label-1>_<scan-type>_<run-number>.mnc
|__ phantom_<subject-1>_<visit-label-1>_<scan-type>_<run-number>.mnc
...
|__ visit_label_2
...
|__ subject_2
|__ subject_3
...
|__ DATS.json
|__ README.md
The ID of the human phantom subject is 963271 and the 9 other subjects present in the dataset are lego phantoms.
Visit labels for the human phantom acquisitions are labelled according to the convention <SiteAbbreviation>_<ScanSessionNumber>_<AcquisitionDate>
Example: MNI_SCAN1_20101125 corresponds to scan session number 1 at the MNI site performed on November 25th, 2010.
The visit labels of the lego phantom acquisitions are labelled according to the convention
<SiteAbbreviation>_<AcquisitionDate>
All images are available in MINC format.
Information about the scanning sites
All MRI scanners were manufacured by SIEMENS.
| Abbreviation | Name | Scanner Model | Scanner software Versions |
|---|---|---|---|
| MNI | Montreal Neurological Institute | TrioTim | Syngo MRI B15 & B17 |
| PH1 | Philadelphia Children's Hospital MR1 | TrioTim | Syngo MRI B15 & B17 |
| PH5 | Philadelphia Children's Hospital MR5 | TrioTim | Syngo MRI B17 |
| SEA | Seattle Children Hospital | TrioTim | Syngo MRI B15 & B17 |
| SLC | St Louis CIR | TrioTim | Syngo MRI B17 |
| SLH | St Louis Children's Hospital | TrioTim | Syngo MRI B15 & B17 |
| SLR | St Louis Washington University | TrioTim | Syngo MRI B15 & B17 |
| UMP | University of Minnesota Prisma Scanner | Prisma fit | Syngo MR D13D |
| UMT | University of Minnesota Tim Trio Scanner | TrioTim | Syngo MR B17 |
| UNH | University of North Carolina Hospital | TrioTim | Syngo MRI B15, B17 & B19 |
| UNR | University of North Carolina Research. | TrioTim | Syngo MR B17 & B19 |
Acknowledgments
The IBIS Network (https://www.ibis-network.org/) and the NIH (https://www.nih.gov/).
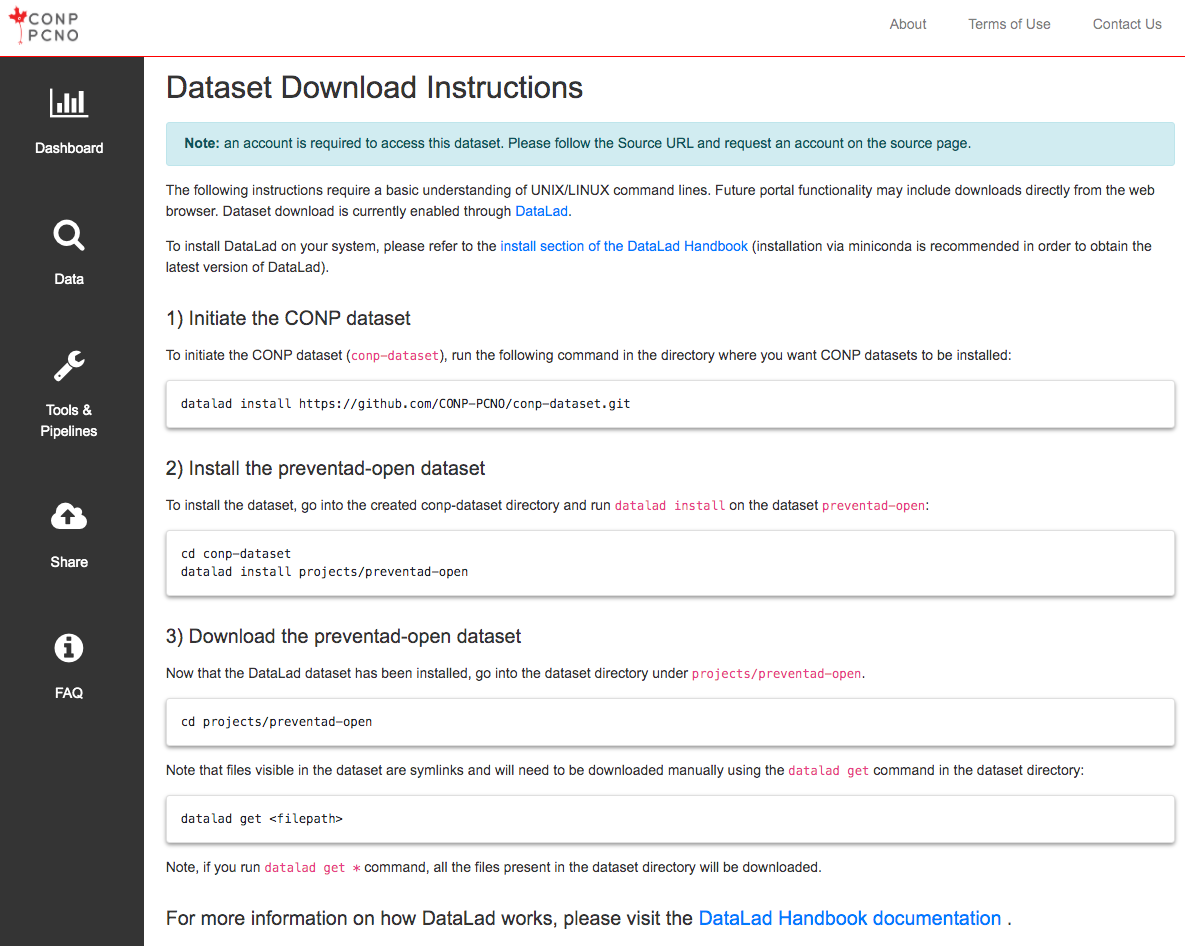
The following instructions require a basic understanding of UNIX/LINUX command lines. A subset of open datasets on the Portal are also available through a browser-based download button. The instructions below regard dataset download with the use of DataLad. To install DataLad on your system, please refer to the install section of the DataLad Handbook .
Note: For maximum compatibility with conp-dataset, the CONP recommends versions 3.12+ of Python, 10.20241202+ of git-annex, and 1.1.4+ of datalad.
1) Initiate the CONP dataset
Run the following command in the directory where you want the CONP dataset (conp-dataset) to be installed:
2) Install the multicenter-phantom dataset
To install the multicenter-phantom dataset, run the following commands to move into the "projects" subdirectory under the "conp-dataset" directory (created in the previous step) and run datalad install:
3) Download data from the multicenter-phantom dataset
Now that the dataset has been installed, go into the multicenter-phantom dataset directory.
The files visible after installing the dataset but before downloading (in the next step) are symbolic links and need to be downloaded manually using the datalad get command:
If you run datalad get * command, all the files available in the dataset directory will be downloaded.
 Experiments - Beta
Experiments - Beta
